Ev ve Ofis taşıma sektöründe lider olmak.Teknolojiyi klrd takip ederek bunu müşteri menuniyeti amacı için kullanmak.Sektörde marka olmak.
İstanbul evden eve nakliyat
Misyonumuz sayesinde edindiğimiz müşteri memnuniyeti ve güven ile müşterilerimizin bizi tavsiye etmelerini sağlamak.
Automated spatio-temporal analysis of dendritic spines and related protein dynamics
.png) This paper presents Dendrite Protein Analysis (DendritePA), a novel automated pattern recognition software to analyze protein trafficking in neurons. Using spatiotemporal information present in multichannel fluorescence videos, the DendritePA generates a temporal maximum intensity projection that enhances the signal-to-noise ratio of important biological structures, segments and tracks dendritic spines, estimates the density of proteins in spines, and analyzes the flux of proteins through the dendrite/spine boundary. Our results also have shown that the activation of cofilin using genetic manipulations leads to immature spines while its inhibition results in an increase in mature spines.
This paper presents Dendrite Protein Analysis (DendritePA), a novel automated pattern recognition software to analyze protein trafficking in neurons. Using spatiotemporal information present in multichannel fluorescence videos, the DendritePA generates a temporal maximum intensity projection that enhances the signal-to-noise ratio of important biological structures, segments and tracks dendritic spines, estimates the density of proteins in spines, and analyzes the flux of proteins through the dendrite/spine boundary. Our results also have shown that the activation of cofilin using genetic manipulations leads to immature spines while its inhibition results in an increase in mature spines.
Spatio-temporal pattern recognition of dendritic spines and protein dynamics using live multichannel fluorescence microscopy
 Actin-regulating proteins, such as cofilin, are
essential in regulating the shape of dendritic spines, and
synaptic plasticity in both neuronal functionality as well as in
neurodegeneration related to aging. Presented is a novel automated pattern recognition system to
analyze protein trafficking in neurons. Using spatio-temporal
information present in multichannel fluorescence videos, the
system generates a temporal maximum intensity projection that
enhances the signal-to-noise ratio of important biological
structures, segments and tracks dendritic spines, and quantifies
the flux and density of proteins in spines. The temporal
dynamics of spines is used to generate spine energy images
which are used to automatically classify the shape of dendritic
spines as stubby, mushroom, or thin. By tracking these spines
over time and using their intensity profiles, the system is able to
analyze the flux patterns of cofilin and other fluorescently
stained proteins.
Actin-regulating proteins, such as cofilin, are
essential in regulating the shape of dendritic spines, and
synaptic plasticity in both neuronal functionality as well as in
neurodegeneration related to aging. Presented is a novel automated pattern recognition system to
analyze protein trafficking in neurons. Using spatio-temporal
information present in multichannel fluorescence videos, the
system generates a temporal maximum intensity projection that
enhances the signal-to-noise ratio of important biological
structures, segments and tracks dendritic spines, and quantifies
the flux and density of proteins in spines. The temporal
dynamics of spines is used to generate spine energy images
which are used to automatically classify the shape of dendritic
spines as stubby, mushroom, or thin. By tracking these spines
over time and using their intensity profiles, the system is able to
analyze the flux patterns of cofilin and other fluorescently
stained proteins.
Dynamic Low-Level Context for the Detection of Mild Traumatic Brain Injury
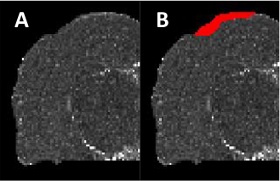 Mild traumatic brain injury (mTBI) appears as low contrast lesions in magnetic resonance (MR) imaging.
Standard automated detection approaches cannot detect the subtle changes caused by the lesions.
We have are proposed and integrated new context features to improve the detection of mTBI lesions.
The approach is validated on a temporal mTBI rat model dataset and shown to have improved dice score and
convergence compared to other state-of-the-art approaches.
Mild traumatic brain injury (mTBI) appears as low contrast lesions in magnetic resonance (MR) imaging.
Standard automated detection approaches cannot detect the subtle changes caused by the lesions.
We have are proposed and integrated new context features to improve the detection of mTBI lesions.
The approach is validated on a temporal mTBI rat model dataset and shown to have improved dice score and
convergence compared to other state-of-the-art approaches.
Automated Detection of Brain Abnormalities in Neonatal Hypoxiaischemic Injury
 We compared the efficacy of three automated brain injury detection methods:
SIRG, HRS, and MWS in human and animal MRI datasets for the detection of
hypoxic ischemic injuries (HIIs). Sensitivity, specificity, and similarity were used as
performance metrics based on manual ("gold standard") injury detection to quantify comparisons.
When compared to the gold standard results SIRG performed the best in 62% of the data,
while results from HRS performed best in 29% of the data, and results for MWS performed the best in 9% of the data.
Injury severity detection revealed that SIRG performed
the best in 67% of the cases, while HRS performed the best in only 33% of the cases. Among these methods, SIRG performs the best in
detecting lesion volumes; HRS is the most robust, while MWS is inferior in both respects.
We compared the efficacy of three automated brain injury detection methods:
SIRG, HRS, and MWS in human and animal MRI datasets for the detection of
hypoxic ischemic injuries (HIIs). Sensitivity, specificity, and similarity were used as
performance metrics based on manual ("gold standard") injury detection to quantify comparisons.
When compared to the gold standard results SIRG performed the best in 62% of the data,
while results from HRS performed best in 29% of the data, and results for MWS performed the best in 9% of the data.
Injury severity detection revealed that SIRG performed
the best in 67% of the cases, while HRS performed the best in only 33% of the cases. Among these methods, SIRG performs the best in
detecting lesion volumes; HRS is the most robust, while MWS is inferior in both respects.
Visual and Contextual Modeling for the Detection of Repeated m-TBI
 Currently, there is a lack of computational methods for the evaluation of mild traumatic brain injury
(mTBI). Furthermore, the development of automated analyses has
been hindered by the subtle nature of mTBI abnormalities.
This research proposes an approach that is able to detect mTBI lesions by combining both the high-level context
and low-level visual information. The visual model utilizes
texture features in MRI along with a probabilistic support vector machine.
Clinically, our approach has the potential to benefit both clinicians by speeding diagnosis and
patients by improving clinical care.
Currently, there is a lack of computational methods for the evaluation of mild traumatic brain injury
(mTBI). Furthermore, the development of automated analyses has
been hindered by the subtle nature of mTBI abnormalities.
This research proposes an approach that is able to detect mTBI lesions by combining both the high-level context
and low-level visual information. The visual model utilizes
texture features in MRI along with a probabilistic support vector machine.
Clinically, our approach has the potential to benefit both clinicians by speeding diagnosis and
patients by improving clinical care.
Detecting Mild Traumatic Brain Injury using Dynamic Low Level Context
 Mild traumatic brain injury is difficult to detect in standard
magnetic resonance (MR) images due to the low contrast
appearance of lesions. In this research a discriminative
approach is presented using a classifier to directly
estimate the posterior probability of lesion at every voxel
using low-level context learned from previous classifiers.
Both visual features including multiple texture measures,
and context features, which include novel features such as
proximity, directional distance, and posterior marginal edge
distance, are used. The context is also taken from previous
time points, so the system automatically captures the
dynamics of the injury progression. The approach is tested
on an mTBI rat model using MR imaging at multiple time
points. Our results show an improved performance in both
the dice score and convergence rate compared to other
approaches.
Mild traumatic brain injury is difficult to detect in standard
magnetic resonance (MR) images due to the low contrast
appearance of lesions. In this research a discriminative
approach is presented using a classifier to directly
estimate the posterior probability of lesion at every voxel
using low-level context learned from previous classifiers.
Both visual features including multiple texture measures,
and context features, which include novel features such as
proximity, directional distance, and posterior marginal edge
distance, are used. The context is also taken from previous
time points, so the system automatically captures the
dynamics of the injury progression. The approach is tested
on an mTBI rat model using MR imaging at multiple time
points. Our results show an improved performance in both
the dice score and convergence rate compared to other
approaches.
Computational Analysis Reveals Increased Blood Deposition
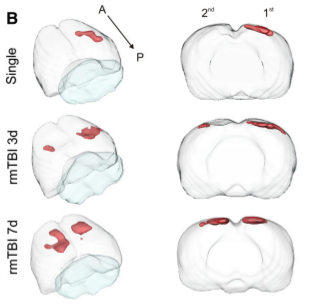 Mild traumatic brain injury (mTBI) has become an increasing public health concern. This study investigated the
temporal development of cortical lesions. Examination of lesion
volume 1d post last injury revealed increased tissue abnormalities within animals compared to other
groups. Histological measurements revealed spatial overlap of regions containing blood
deposition and microglial activation within the cortices of all animals. Our findings suggest that
there is a window of tissue vulnerability where a second distant mTBI, induced 7d after an initial injury,
exacerbates tissue abnormalities consistent with hemorrhagic progression.
Mild traumatic brain injury (mTBI) has become an increasing public health concern. This study investigated the
temporal development of cortical lesions. Examination of lesion
volume 1d post last injury revealed increased tissue abnormalities within animals compared to other
groups. Histological measurements revealed spatial overlap of regions containing blood
deposition and microglial activation within the cortices of all animals. Our findings suggest that
there is a window of tissue vulnerability where a second distant mTBI, induced 7d after an initial injury,
exacerbates tissue abnormalities consistent with hemorrhagic progression.
Contextual and Visual Modeling for Detection of Mild Traumatic Brain Injury in MRI
 Mild traumatic brain injury (mTBI) is difficult to detect as
the current tools are qualitative, which can lead to poor
diagnosis and treatment. The low contrast appearance of
mTBI abnormalities on magnetic resonance (MR) images
makes quantification problematic for image processing and
analysis techniques. To overcome these difficulties, an
algorithm is proposed that takes advantage of subject
information and texture information from MR images. A
contextual model is developed to simulate the progression of
the disease using multiple inputs, such as the time postinjury
and the location of injury. Textural features are used
along with feature selection for a single MR modality.
Results from a probabilistic support vector machine using
textural features are fused with the contextual model to
obtain a robust estimation of abnormal tissue. A novel rat
temporal dataset demonstrates the ability of our approach
to outperform other state of the art approaches.
Mild traumatic brain injury (mTBI) is difficult to detect as
the current tools are qualitative, which can lead to poor
diagnosis and treatment. The low contrast appearance of
mTBI abnormalities on magnetic resonance (MR) images
makes quantification problematic for image processing and
analysis techniques. To overcome these difficulties, an
algorithm is proposed that takes advantage of subject
information and texture information from MR images. A
contextual model is developed to simulate the progression of
the disease using multiple inputs, such as the time postinjury
and the location of injury. Textural features are used
along with feature selection for a single MR modality.
Results from a probabilistic support vector machine using
textural features are fused with the contextual model to
obtain a robust estimation of abnormal tissue. A novel rat
temporal dataset demonstrates the ability of our approach
to outperform other state of the art approaches.
Computational Analysis of Injured Tissue Following Repetitive Mild Traumatic Brain Injury
.png) Mild traumatic brain injury (mTBI) has become an increasing public health concern as subsequent injuries can exacerbate existing neuropathology and result in neurological deficits. Experimental models of repetitive mTBI have explored injuries induced to the same location; however our study focuses on tissue level alterations following two contralateral mTBIs.
Mild traumatic brain injury (mTBI) has become an increasing public health concern as subsequent injuries can exacerbate existing neuropathology and result in neurological deficits. Experimental models of repetitive mTBI have explored injuries induced to the same location; however our study focuses on tissue level alterations following two contralateral mTBIs.
Mild traumatic brain injury detection through visual and contextual modeling
.png) This paper proposes an approach that is able to detect mTBI lesions by combining both the high-level context and low-level visual information. The contextual model estimates the progression of the disease using subject information, such as the time since injury and the knowledge about the location of mTBI. The visual model utilizes texture features in MRI along with a probabilistic support vector machine to maximize the discrimination in unimodal MR images. These two models are fused to obtain a final estimate of the locations of the mTBI lesion. The models are tested using a novel rodent model of repeated mTBI dataset. The experimental results demonstrate that the fusion of both contextual and visual textural features outperforms other state-of-the-art approaches. Clinically, our approach has the potential to benefit both clinicians by speeding diagnosis and patients by improving clinical care.
This paper proposes an approach that is able to detect mTBI lesions by combining both the high-level context and low-level visual information. The contextual model estimates the progression of the disease using subject information, such as the time since injury and the knowledge about the location of mTBI. The visual model utilizes texture features in MRI along with a probabilistic support vector machine to maximize the discrimination in unimodal MR images. These two models are fused to obtain a final estimate of the locations of the mTBI lesion. The models are tested using a novel rodent model of repeated mTBI dataset. The experimental results demonstrate that the fusion of both contextual and visual textural features outperforms other state-of-the-art approaches. Clinically, our approach has the potential to benefit both clinicians by speeding diagnosis and patients by improving clinical care.
Automated Ischemic Lesion Detection in a NeonatalModel of Hypoxic Ischemic Injury
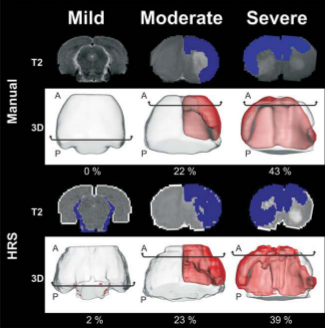 Here we introduce and compare an automated detection system for ischemic lesions in a neonatal
model of BCAO-H from T2 weighted MRI (T2WI) to
the currently used "gold standard" of manual segmentation. Forty-three, P10 BCAO-H rat pups and
8 controls underwent T2WI for 1 day and for 28 days. A computational imaging method - Hierarchical Region
Splitting (HRS) - was developed to automatically and rapidly detect and quantify 3D lesion and
normal appearing brain matter (NABM) volumes. HRS quantified lesions and NABM volumes within 15 seconds in
comparison to 3 hours for its manual counterpart.
Here we introduce and compare an automated detection system for ischemic lesions in a neonatal
model of BCAO-H from T2 weighted MRI (T2WI) to
the currently used "gold standard" of manual segmentation. Forty-three, P10 BCAO-H rat pups and
8 controls underwent T2WI for 1 day and for 28 days. A computational imaging method - Hierarchical Region
Splitting (HRS) - was developed to automatically and rapidly detect and quantify 3D lesion and
normal appearing brain matter (NABM) volumes. HRS quantified lesions and NABM volumes within 15 seconds in
comparison to 3 hours for its manual counterpart.
Semi-Automated Segmentation of ADC Maps Reliably Defines Ischemic Perinatal Stroke Injury
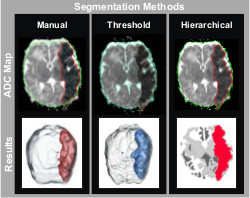 Ischemic brain tissue has a signature hypointensity on ADC maps that indicates
a net reduction in water diffusion relative to normal unaffected tissue. The
ischemic lesions induced by embolic arterial stroke are prominent within the
first 72 hrs of injury and can be defined by identifying and segmenting the
hypointense region. Assessment of these ischemic lesions soon after injury
would help clinicians make timely, informative treatment decisions. Manual volumetric injury quantification is not used
clinically because it is a labor intensive process. Observer (inter-/intra-)
variability can also be problematic; however, it remains the gold standard. We
used a multi-disciplinary approach to develop two semi-automated methods of
ischemic injury segmentation: ADC thresholding and hierarchical region
splitting, and compared them with manual segmentation results. We believe
that this multi-disciplinary approach will provide a fundamental basis for the
development of novel and clinically relevant automated segmentation software for rapid and unbiased ischemic injury
discrimination.
Ischemic brain tissue has a signature hypointensity on ADC maps that indicates
a net reduction in water diffusion relative to normal unaffected tissue. The
ischemic lesions induced by embolic arterial stroke are prominent within the
first 72 hrs of injury and can be defined by identifying and segmenting the
hypointense region. Assessment of these ischemic lesions soon after injury
would help clinicians make timely, informative treatment decisions. Manual volumetric injury quantification is not used
clinically because it is a labor intensive process. Observer (inter-/intra-)
variability can also be problematic; however, it remains the gold standard. We
used a multi-disciplinary approach to develop two semi-automated methods of
ischemic injury segmentation: ADC thresholding and hierarchical region
splitting, and compared them with manual segmentation results. We believe
that this multi-disciplinary approach will provide a fundamental basis for the
development of novel and clinically relevant automated segmentation software for rapid and unbiased ischemic injury
discrimination.
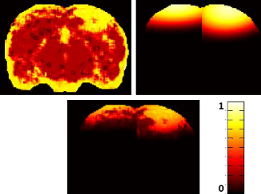
Symmetry-Integrated Injury Detection for Brain MRI
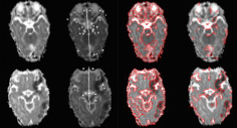 We present a new brain injury detection approach in images acquired by magnetic resonance
imaging (MRI). The proposed approach is based on the fact that the anatomical structure of a 2D brain is
highly symmetric, while most of the injury in the brain generally indicates asymmetry. The approach starts
from symmetry integrated region growing segmentation of the brain images using the symmetry affinity
matrix, and candidate asymmetric regions are initially extracted using kurtosis and skewness of symmetry
affinity matrix. An Expectation Maximum classifier with Gaussian mixture model is used explicitly to classify
asymmetric regions into two sections: injury and non-injury. Experimental results are carried out to demonstrate the
efficacy of the approach for injury detection.
We present a new brain injury detection approach in images acquired by magnetic resonance
imaging (MRI). The proposed approach is based on the fact that the anatomical structure of a 2D brain is
highly symmetric, while most of the injury in the brain generally indicates asymmetry. The approach starts
from symmetry integrated region growing segmentation of the brain images using the symmetry affinity
matrix, and candidate asymmetric regions are initially extracted using kurtosis and skewness of symmetry
affinity matrix. An Expectation Maximum classifier with Gaussian mixture model is used explicitly to classify
asymmetric regions into two sections: injury and non-injury. Experimental results are carried out to demonstrate the
efficacy of the approach for injury detection.
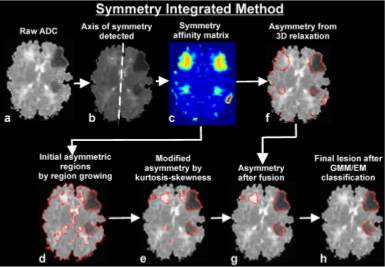
Automatic Symmetry-Integrated Brain Injury Detection in MRI Sequences
.png) This paper presents a fully automated symmetry-integrated brain injury detection method for magnetic resonance imaging (MRI) sequences. One of the limitations of current injury detection methods often involves a large amount of training data or a prior model that is only applicable to a limited domain of brain slices, with low computational efficiency and robustness. Our proposed approach can detect injuries from a wide variety of brain images since it makes use of symmetry as a dominant feature, and does not rely on any prior models and training phases. The approach consists of the following steps: (a) symmetry integrated segmentation of brain slices based on symmetry affinity matrix, (b) computation of kurtosis and skewness of symmetry affinity matrix to find potential asymmetric regions, (c) clustering of the pixels in symmetry affinity matrix using a 3D relaxation algorithm, (d) fusion of the results of (b) and (c) to obtain refined asymmetric regions, (e) Gaussian mixture model for unsupervised classification of potential asymmetric regions as the set of regions corresponding to brain injuries. Experimental results are carried out to demonstrate the efficacy of the approach.
This paper presents a fully automated symmetry-integrated brain injury detection method for magnetic resonance imaging (MRI) sequences. One of the limitations of current injury detection methods often involves a large amount of training data or a prior model that is only applicable to a limited domain of brain slices, with low computational efficiency and robustness. Our proposed approach can detect injuries from a wide variety of brain images since it makes use of symmetry as a dominant feature, and does not rely on any prior models and training phases. The approach consists of the following steps: (a) symmetry integrated segmentation of brain slices based on symmetry affinity matrix, (b) computation of kurtosis and skewness of symmetry affinity matrix to find potential asymmetric regions, (c) clustering of the pixels in symmetry affinity matrix using a 3D relaxation algorithm, (d) fusion of the results of (b) and (c) to obtain refined asymmetric regions, (e) Gaussian mixture model for unsupervised classification of potential asymmetric regions as the set of regions corresponding to brain injuries. Experimental results are carried out to demonstrate the efficacy of the approach.
|


.png) This paper presents Dendrite Protein Analysis (DendritePA), a novel automated pattern recognition software to analyze protein trafficking in neurons. Using spatiotemporal information present in multichannel fluorescence videos, the DendritePA generates a temporal maximum intensity projection that enhances the signal-to-noise ratio of important biological structures, segments and tracks dendritic spines, estimates the density of proteins in spines, and analyzes the flux of proteins through the dendrite/spine boundary. Our results also have shown that the activation of cofilin using genetic manipulations leads to immature spines while its inhibition results in an increase in mature spines.
This paper presents Dendrite Protein Analysis (DendritePA), a novel automated pattern recognition software to analyze protein trafficking in neurons. Using spatiotemporal information present in multichannel fluorescence videos, the DendritePA generates a temporal maximum intensity projection that enhances the signal-to-noise ratio of important biological structures, segments and tracks dendritic spines, estimates the density of proteins in spines, and analyzes the flux of proteins through the dendrite/spine boundary. Our results also have shown that the activation of cofilin using genetic manipulations leads to immature spines while its inhibition results in an increase in mature spines.
 Mild traumatic brain injury (mTBI) appears as low contrast lesions in magnetic resonance (MR) imaging.
Standard automated detection approaches cannot detect the subtle changes caused by the lesions.
We have are proposed and integrated new context features to improve the detection of mTBI lesions.
The approach is validated on a temporal mTBI rat model dataset and shown to have improved dice score and
convergence compared to other state-of-the-art approaches.
Mild traumatic brain injury (mTBI) appears as low contrast lesions in magnetic resonance (MR) imaging.
Standard automated detection approaches cannot detect the subtle changes caused by the lesions.
We have are proposed and integrated new context features to improve the detection of mTBI lesions.
The approach is validated on a temporal mTBI rat model dataset and shown to have improved dice score and
convergence compared to other state-of-the-art approaches.
 We compared the efficacy of three automated brain injury detection methods:
SIRG, HRS, and MWS in human and animal MRI datasets for the detection of
hypoxic ischemic injuries (HIIs). Sensitivity, specificity, and similarity were used as
performance metrics based on manual ("gold standard") injury detection to quantify comparisons.
When compared to the gold standard results SIRG performed the best in 62% of the data,
while results from HRS performed best in 29% of the data, and results for MWS performed the best in 9% of the data.
Injury severity detection revealed that SIRG performed
the best in 67% of the cases, while HRS performed the best in only 33% of the cases. Among these methods, SIRG performs the best in
detecting lesion volumes; HRS is the most robust, while MWS is inferior in both respects.
We compared the efficacy of three automated brain injury detection methods:
SIRG, HRS, and MWS in human and animal MRI datasets for the detection of
hypoxic ischemic injuries (HIIs). Sensitivity, specificity, and similarity were used as
performance metrics based on manual ("gold standard") injury detection to quantify comparisons.
When compared to the gold standard results SIRG performed the best in 62% of the data,
while results from HRS performed best in 29% of the data, and results for MWS performed the best in 9% of the data.
Injury severity detection revealed that SIRG performed
the best in 67% of the cases, while HRS performed the best in only 33% of the cases. Among these methods, SIRG performs the best in
detecting lesion volumes; HRS is the most robust, while MWS is inferior in both respects.
 Currently, there is a lack of computational methods for the evaluation of mild traumatic brain injury
(mTBI). Furthermore, the development of automated analyses has
been hindered by the subtle nature of mTBI abnormalities.
This research proposes an approach that is able to detect mTBI lesions by combining both the high-level context
and low-level visual information. The visual model utilizes
texture features in MRI along with a probabilistic support vector machine.
Clinically, our approach has the potential to benefit both clinicians by speeding diagnosis and
patients by improving clinical care.
Currently, there is a lack of computational methods for the evaluation of mild traumatic brain injury
(mTBI). Furthermore, the development of automated analyses has
been hindered by the subtle nature of mTBI abnormalities.
This research proposes an approach that is able to detect mTBI lesions by combining both the high-level context
and low-level visual information. The visual model utilizes
texture features in MRI along with a probabilistic support vector machine.
Clinically, our approach has the potential to benefit both clinicians by speeding diagnosis and
patients by improving clinical care.
 Mild traumatic brain injury is difficult to detect in standard
magnetic resonance (MR) images due to the low contrast
appearance of lesions. In this research a discriminative
approach is presented using a classifier to directly
estimate the posterior probability of lesion at every voxel
using low-level context learned from previous classifiers.
Both visual features including multiple texture measures,
and context features, which include novel features such as
proximity, directional distance, and posterior marginal edge
distance, are used. The context is also taken from previous
time points, so the system automatically captures the
dynamics of the injury progression. The approach is tested
on an mTBI rat model using MR imaging at multiple time
points. Our results show an improved performance in both
the dice score and convergence rate compared to other
approaches.
Mild traumatic brain injury is difficult to detect in standard
magnetic resonance (MR) images due to the low contrast
appearance of lesions. In this research a discriminative
approach is presented using a classifier to directly
estimate the posterior probability of lesion at every voxel
using low-level context learned from previous classifiers.
Both visual features including multiple texture measures,
and context features, which include novel features such as
proximity, directional distance, and posterior marginal edge
distance, are used. The context is also taken from previous
time points, so the system automatically captures the
dynamics of the injury progression. The approach is tested
on an mTBI rat model using MR imaging at multiple time
points. Our results show an improved performance in both
the dice score and convergence rate compared to other
approaches.
 Mild traumatic brain injury (mTBI) has become an increasing public health concern. This study investigated the
temporal development of cortical lesions. Examination of lesion
volume 1d post last injury revealed increased tissue abnormalities within animals compared to other
groups. Histological measurements revealed spatial overlap of regions containing blood
deposition and microglial activation within the cortices of all animals. Our findings suggest that
there is a window of tissue vulnerability where a second distant mTBI, induced 7d after an initial injury,
exacerbates tissue abnormalities consistent with hemorrhagic progression.
Mild traumatic brain injury (mTBI) has become an increasing public health concern. This study investigated the
temporal development of cortical lesions. Examination of lesion
volume 1d post last injury revealed increased tissue abnormalities within animals compared to other
groups. Histological measurements revealed spatial overlap of regions containing blood
deposition and microglial activation within the cortices of all animals. Our findings suggest that
there is a window of tissue vulnerability where a second distant mTBI, induced 7d after an initial injury,
exacerbates tissue abnormalities consistent with hemorrhagic progression.
 Mild traumatic brain injury (mTBI) is difficult to detect as
the current tools are qualitative, which can lead to poor
diagnosis and treatment. The low contrast appearance of
mTBI abnormalities on magnetic resonance (MR) images
makes quantification problematic for image processing and
analysis techniques. To overcome these difficulties, an
algorithm is proposed that takes advantage of subject
information and texture information from MR images. A
contextual model is developed to simulate the progression of
the disease using multiple inputs, such as the time postinjury
and the location of injury. Textural features are used
along with feature selection for a single MR modality.
Results from a probabilistic support vector machine using
textural features are fused with the contextual model to
obtain a robust estimation of abnormal tissue. A novel rat
temporal dataset demonstrates the ability of our approach
to outperform other state of the art approaches.
Mild traumatic brain injury (mTBI) is difficult to detect as
the current tools are qualitative, which can lead to poor
diagnosis and treatment. The low contrast appearance of
mTBI abnormalities on magnetic resonance (MR) images
makes quantification problematic for image processing and
analysis techniques. To overcome these difficulties, an
algorithm is proposed that takes advantage of subject
information and texture information from MR images. A
contextual model is developed to simulate the progression of
the disease using multiple inputs, such as the time postinjury
and the location of injury. Textural features are used
along with feature selection for a single MR modality.
Results from a probabilistic support vector machine using
textural features are fused with the contextual model to
obtain a robust estimation of abnormal tissue. A novel rat
temporal dataset demonstrates the ability of our approach
to outperform other state of the art approaches.
.png) Mild traumatic brain injury (mTBI) has become an increasing public health concern as subsequent injuries can exacerbate existing neuropathology and result in neurological deficits. Experimental models of repetitive mTBI have explored injuries induced to the same location; however our study focuses on tissue level alterations following two contralateral mTBIs.
Mild traumatic brain injury (mTBI) has become an increasing public health concern as subsequent injuries can exacerbate existing neuropathology and result in neurological deficits. Experimental models of repetitive mTBI have explored injuries induced to the same location; however our study focuses on tissue level alterations following two contralateral mTBIs.
.png) This paper proposes an approach that is able to detect mTBI lesions by combining both the high-level context and low-level visual information. The contextual model estimates the progression of the disease using subject information, such as the time since injury and the knowledge about the location of mTBI. The visual model utilizes texture features in MRI along with a probabilistic support vector machine to maximize the discrimination in unimodal MR images. These two models are fused to obtain a final estimate of the locations of the mTBI lesion. The models are tested using a novel rodent model of repeated mTBI dataset. The experimental results demonstrate that the fusion of both contextual and visual textural features outperforms other state-of-the-art approaches. Clinically, our approach has the potential to benefit both clinicians by speeding diagnosis and patients by improving clinical care.
This paper proposes an approach that is able to detect mTBI lesions by combining both the high-level context and low-level visual information. The contextual model estimates the progression of the disease using subject information, such as the time since injury and the knowledge about the location of mTBI. The visual model utilizes texture features in MRI along with a probabilistic support vector machine to maximize the discrimination in unimodal MR images. These two models are fused to obtain a final estimate of the locations of the mTBI lesion. The models are tested using a novel rodent model of repeated mTBI dataset. The experimental results demonstrate that the fusion of both contextual and visual textural features outperforms other state-of-the-art approaches. Clinically, our approach has the potential to benefit both clinicians by speeding diagnosis and patients by improving clinical care.
 Here we introduce and compare an automated detection system for ischemic lesions in a neonatal
model of BCAO-H from T2 weighted MRI (T2WI) to
the currently used "gold standard" of manual segmentation. Forty-three, P10 BCAO-H rat pups and
8 controls underwent T2WI for 1 day and for 28 days. A computational imaging method - Hierarchical Region
Splitting (HRS) - was developed to automatically and rapidly detect and quantify 3D lesion and
normal appearing brain matter (NABM) volumes. HRS quantified lesions and NABM volumes within 15 seconds in
comparison to 3 hours for its manual counterpart.
Here we introduce and compare an automated detection system for ischemic lesions in a neonatal
model of BCAO-H from T2 weighted MRI (T2WI) to
the currently used "gold standard" of manual segmentation. Forty-three, P10 BCAO-H rat pups and
8 controls underwent T2WI for 1 day and for 28 days. A computational imaging method - Hierarchical Region
Splitting (HRS) - was developed to automatically and rapidly detect and quantify 3D lesion and
normal appearing brain matter (NABM) volumes. HRS quantified lesions and NABM volumes within 15 seconds in
comparison to 3 hours for its manual counterpart.
 Ischemic brain tissue has a signature hypointensity on ADC maps that indicates
a net reduction in water diffusion relative to normal unaffected tissue. The
ischemic lesions induced by embolic arterial stroke are prominent within the
first 72 hrs of injury and can be defined by identifying and segmenting the
hypointense region. Assessment of these ischemic lesions soon after injury
would help clinicians make timely, informative treatment decisions. Manual volumetric injury quantification is not used
clinically because it is a labor intensive process. Observer (inter-/intra-)
variability can also be problematic; however, it remains the gold standard. We
used a multi-disciplinary approach to develop two semi-automated methods of
ischemic injury segmentation: ADC thresholding and hierarchical region
splitting, and compared them with manual segmentation results. We believe
that this multi-disciplinary approach will provide a fundamental basis for the
development of novel and clinically relevant automated segmentation software for rapid and unbiased ischemic injury
discrimination.
Ischemic brain tissue has a signature hypointensity on ADC maps that indicates
a net reduction in water diffusion relative to normal unaffected tissue. The
ischemic lesions induced by embolic arterial stroke are prominent within the
first 72 hrs of injury and can be defined by identifying and segmenting the
hypointense region. Assessment of these ischemic lesions soon after injury
would help clinicians make timely, informative treatment decisions. Manual volumetric injury quantification is not used
clinically because it is a labor intensive process. Observer (inter-/intra-)
variability can also be problematic; however, it remains the gold standard. We
used a multi-disciplinary approach to develop two semi-automated methods of
ischemic injury segmentation: ADC thresholding and hierarchical region
splitting, and compared them with manual segmentation results. We believe
that this multi-disciplinary approach will provide a fundamental basis for the
development of novel and clinically relevant automated segmentation software for rapid and unbiased ischemic injury
discrimination.
 We present a new brain injury detection approach in images acquired by magnetic resonance
imaging (MRI). The proposed approach is based on the fact that the anatomical structure of a 2D brain is
highly symmetric, while most of the injury in the brain generally indicates asymmetry. The approach starts
from symmetry integrated region growing segmentation of the brain images using the symmetry affinity
matrix, and candidate asymmetric regions are initially extracted using kurtosis and skewness of symmetry
affinity matrix. An Expectation Maximum classifier with Gaussian mixture model is used explicitly to classify
asymmetric regions into two sections: injury and non-injury. Experimental results are carried out to demonstrate the
efficacy of the approach for injury detection.
We present a new brain injury detection approach in images acquired by magnetic resonance
imaging (MRI). The proposed approach is based on the fact that the anatomical structure of a 2D brain is
highly symmetric, while most of the injury in the brain generally indicates asymmetry. The approach starts
from symmetry integrated region growing segmentation of the brain images using the symmetry affinity
matrix, and candidate asymmetric regions are initially extracted using kurtosis and skewness of symmetry
affinity matrix. An Expectation Maximum classifier with Gaussian mixture model is used explicitly to classify
asymmetric regions into two sections: injury and non-injury. Experimental results are carried out to demonstrate the
efficacy of the approach for injury detection.
.png) This paper presents a fully automated symmetry-integrated brain injury detection method for magnetic resonance imaging (MRI) sequences. One of the limitations of current injury detection methods often involves a large amount of training data or a prior model that is only applicable to a limited domain of brain slices, with low computational efficiency and robustness. Our proposed approach can detect injuries from a wide variety of brain images since it makes use of symmetry as a dominant feature, and does not rely on any prior models and training phases. The approach consists of the following steps: (a) symmetry integrated segmentation of brain slices based on symmetry affinity matrix, (b) computation of kurtosis and skewness of symmetry affinity matrix to find potential asymmetric regions, (c) clustering of the pixels in symmetry affinity matrix using a 3D relaxation algorithm, (d) fusion of the results of (b) and (c) to obtain refined asymmetric regions, (e) Gaussian mixture model for unsupervised classification of potential asymmetric regions as the set of regions corresponding to brain injuries. Experimental results are carried out to demonstrate the efficacy of the approach.
This paper presents a fully automated symmetry-integrated brain injury detection method for magnetic resonance imaging (MRI) sequences. One of the limitations of current injury detection methods often involves a large amount of training data or a prior model that is only applicable to a limited domain of brain slices, with low computational efficiency and robustness. Our proposed approach can detect injuries from a wide variety of brain images since it makes use of symmetry as a dominant feature, and does not rely on any prior models and training phases. The approach consists of the following steps: (a) symmetry integrated segmentation of brain slices based on symmetry affinity matrix, (b) computation of kurtosis and skewness of symmetry affinity matrix to find potential asymmetric regions, (c) clustering of the pixels in symmetry affinity matrix using a 3D relaxation algorithm, (d) fusion of the results of (b) and (c) to obtain refined asymmetric regions, (e) Gaussian mixture model for unsupervised classification of potential asymmetric regions as the set of regions corresponding to brain injuries. Experimental results are carried out to demonstrate the efficacy of the approach.